Bacterial Communities in Plastic-Contaminated Sites and Rusted Acrylic Wall Paint: Isolation and Characterization
| Received 24 Apr, 2023 |
Accepted 26 Nov, 2023 |
Published 01 Jan, 2024 |
Background and Objective: The possibility of microbial organisms exist in polluted environmental conditions, including long-term plastic waste landfills and also over the rusted walls at the water leak pipe regions. The present study was carried out in a major plastic dumpsite and rusted acrylic wall paint and was conceded to study the bacterial community from the contaminated sites. Materials and Methods: Soil samples were collected separately from a plastic waste filled area, for a longer period and from the rusting zone due to sanitary pipe leakage on a building's exterior wall. Serial dilution, direct isolation and pure culture methods were employed to isolate Bacterial species from those experimental samples and further identified the bacterial groups, using standard manuals. Results: A total of eighteen bacterial species were isolated and identified which include Corynebacterium sp., Flavobacterium sp., Neisseria sp., Staphylococcus sp., Streptococcus sp., Pseudomonas sp., Bacillus sp., Klebsiella sp., Bacillus sp., Corynebacterium sp., Pseudomonas sp., Escherichia coli, Bacillus sp., Yersinia sp., Planococcus sp., Yersinia sp., Klebsiella sp. and Agrobacterium sp. Conclusion: The abundant nature of bacterial species existence was noticed in the soil samples collected from the long-term land filled area with plastic wastes and in the rusted exterior wall scrap. The potential nature of the bacteria species isolated from this experiment could be applied further to test their plastic degradation potential.
| Copyright © 2024 Subramanian et al. This is an open-access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |
INTRODUCTION
Plastic wastes make up about 10% of household garbage and are primarily disposed of in landfills1 and causing land and soil contamination and also this effect extends to pollute the aquatic system can then spread to the aquatic environment2. Even though landfilling is the most widely used and this ease of the waste management strategy has been widely followed, mainly due to the unavailability of land area1; however, the after effects on the ecological conditions of the land and water environments become much worse than due to the landfills of other domestic wastes. Monitoring of these sites became more difficult having a wider physical and chemical impact on the environment and human health. If landfills are properly managed and other strategies including reduction, recycling and reusing this menace of the deterioration of the environment and dangers to public health can be minimized. Disintegrating plastic residues is the option of preventing soil pollution by plastic wastes to some extent has also been emphasized3.
The microbes that develop on plastics are distinct from those in the nearby water, soil and organic particles4,5. The selection of microbial communities at the start of colonization is greatly influenced by the nature of plastic surfaces6. The establishment of microbial communities on the polymers can influence some pioneer novel bacteria7. The combined effects of geographic regions8 and the kind of polymer have an impact on the microorganism that proliferates on plastics9.
Microorganisms encountered in contaminated environments have been identified and characterized through traditional culture-dependent approaches10. Extreme environments are too many and unexplored for the occurrence of microbes as extremophiles11 and their biological characterization is required to be carried out. Many normal and extreme bacterial species have been isolated and utilized as novel biodegrading agents; for instance, degrading petroleum hydrocarbons12, also found to have a vital degradation role in synthetic polymer13-17. Algae and cyanobacteria are found to have deteriorating nature and thrive better over light-exposed surfaces in buildings18-20, they’re predisposing the surface to colonization by potentially more damaging organisms such as fungi, mosses and higher plants21.
Microbes cause cleavage of the polymer chain using certain enzymes and convert them into monomers and oligomers. The diverse metabolic capability of microbes can be exploited for the bioremediation of plastic wastes that use microbial strain developed through selection and strain improvement21. Most of the available research demonstrated the possibility of soil microbial isolates have the potential to degrade plastics; however, a clear-cut understanding of the microbial community that exists in the rusted wall, coated using acrylic paints and also in the long-term plastic waste landfills is necessary to be developed. Such research results would be useful in the contribution of scientific knowledge on the specific microbes dwelling in those contaminated environments, which would be further can be applied in the testing of the biodegrading ability of plastic wastes. In this experiment, an attempt was made to isolate the bacterial community from a long-term plastic landfill site and from the rusted wall, coated with acrylic wall paint considering it as the initial step to mitigate plastic pollution through effective degradation by isolated novel bacterial species.
MATERIALS AND METHODS
Sampling site description: The experiment was carried out in Thiagarajar College, Madurai, (9.8821°N, 78.0816°E; 134/439.63 feet m2) southern India. Semi-arid ecoclimatic condition is prevalent in this experimental area with an average annual precipitation ranges between 100 to 110 mm. The soil samples used in this study were collected from two different sites, the plastic waste dumped sites in Uthangudi Village at Latitude: 9°57'22"N and Longitude: 78°10'14" E, Madurai, Tamil Nadu. The site was found to have been utilized for about 13 years, as a plastic waste landfill and segregation site. Soil sample was collected from this long-term plastic waste dump site, using the soil auger and the soil sample was collected from 10×10×10 cm size area. The second experimental sample was collected by scrapping the exterior wall at the rusted zones of a 75 year old building in Thiagarajar College at Latitude: 9°54'43"N; Longitude: 78°8'48"E, Madurai, Tamil Nadu rusting of the exterior wall was noticed with the degrading acrylic coat over the wall, due to leakage pipe fittings, laid over the wall.
Sampling: Soil samples were collected at three different randomly select points at the long-term plastic wastes landfill site, using the soil auger of 10x10x10 cm area and the bulk density was recorded with an average value of 0.50 kg cm–3. Soil samples collected from each point was pooled together and stored in a plastic bag for further laboratory analysis. The wall scrap of the rusted wall zone, with the rusted acrylic waste was collected from the four different zones of water leakage points over the water pipe outlets, fitted on the wall surface and a total weight of approximately 100 g of wall scrap sample was obtained. The soil sample from the plastic waste landfill site and from the rusted acrylic material painted wall were collected and stored in sterilized polythene bags and stored at 20°C, for further biochemical and microbial analysis.
Physical nature of collected samples: Physical parameters were analyzed with standard methods including pH, conductivity and salinity using a water analyzer kit (Systronics Make, Model: 371). The acidity and alkalinity were analyzed, using wet chemical analytical methods.
Isolation of bacteria from contaminated sites
Sample processing and dilution: Soil sample collected from the long-term plastic wastes was used to prepare a 10–1 stock solution, using sterile distilled water for making the soil solution. Serial dilutions from a range between 10–2 and 10–5 were prepared using the 10–1 stock solution. From each dilution, a 0.1 mL soil solution sample was inoculated into sterilized petri plates containing solidified nutrient agar medium to isolate the available bacterial organisms. The serially diluted sample was inoculated over the medium using the spread plate method and incubated at 30°C for 24 hrs. After incubation, microbial colonies were observed. From the purely isolated colonies, sub-culturing of the isolated culture was done to obtain the adequate quantity for further experiments, using the standard procedure22. The isolated pure cultures were inoculated onto agar plates and stored.
Identification of microorganisms: Biochemical characterization of the bacterial isolates was carried out using Bergey’s manual23, through which, the bacterial groups were identified. The analytical procedures, viz., Gram staining, morphological evaluation, catalase activity test, carbohydrate fermentation, starch hydrolysis, gas production, motility test, Indole test, H2S evolution test, methyl red test, protease test and voges-proskauer Test (VP) was carried out for the confirmation of the microbial communities, exist in the experimental samples.
RESULTS
The pH values were found slightly higher than the neutral pH, in the samples collected from both experimental sites. The salinity is remarkably higher in the plastic-contaminated soil than in the sample collected from rusted acrylic wall paint. Acidity and alkalinity are quite similar in both experimental sites (Table 1).
The colony morphological features of isolated bacteria were studied discretely and tabulated in Table 2. Nine yellow and eight white colonies and only one colony pink colour colony formation were noted and all colonies except two colonies were observed spreading into spherical shapes which were irregular.
| Table 1: | Physical characteristic features of soil samples, collected from the experimental sites | |||
Physical characters of the soil sample |
||||||||
| Experimental site | Colour |
Texture |
Odor |
pH |
Conductivity |
Salinity |
Acidity |
Alkalinity |
| Long-term plastic waste landfill area |
Brown |
Rough, coarse |
Garbage odor |
7.24 |
228.6 |
0.06 |
0.384 |
0.365 |
| Rusted wall coated with acrylic paint |
Blackish brown |
Rough, flaky |
Odorless |
7.62 |
129 |
0.03 |
0.378 |
0.385 |
| Table 2: | Morphological features of Bacterial isolates from the experimental soils | |||
| Isolate | Size |
Color |
Texture |
Form |
Elevation |
Margin |
| A | Small |
White |
Smooth |
Round |
Raised |
Serrate |
| B | Small |
Yellow |
Smooth |
Round |
Flat |
Entire |
| C | Large |
Pink |
Dry |
Round |
Flat |
Scalloped |
| D | Large |
White |
Smooth |
Irregular |
Raised |
Undulate |
| E | Small |
Yellow |
Viscid |
Round |
Flat |
Entire |
| F | Moderate |
White |
Dry |
Round |
Flat |
Undulate |
| G | Small |
Yellow |
Mucoid |
Round |
Convex |
Entire |
| H | Small |
White |
Mucoid |
Round |
Convex |
Entire |
| I | Small |
White |
Viscid |
Round |
Convex |
Entire |
| J | Small |
Yellow |
Mucoid |
Round |
Convex |
Entire |
| K | Moderate |
White |
Slime |
Irregular |
Convex |
Undulate |
| L | Moderate |
Yellow |
Mucoid |
Round |
Flat |
Entire |
| M | Small |
Pale yellow |
Viscid |
Round |
Flat |
Entire |
| N | Moderate |
White |
Dry |
Round |
Raised |
Entire |
| O | Moderate |
White |
Smooth |
Round |
Flat |
Entire |
| P | Small |
Pale yellow |
Mucus |
Round |
Convex |
Entire |
| Q | Small |
Yellow |
Smooth |
Round |
Flat |
Entire |
| R | Small |
Yellow |
Smooth |
Round |
Flat |
Entire |
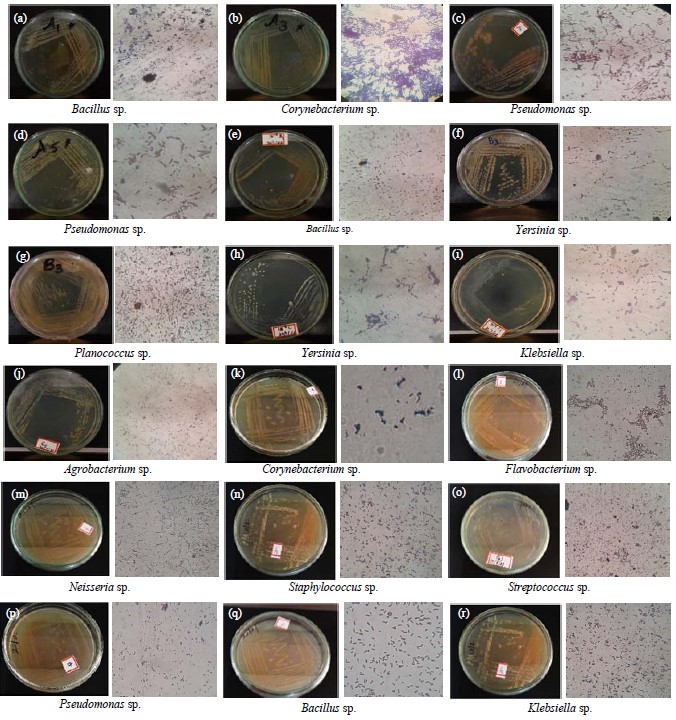
Figure 1(a-r) portrayed with the following bacterial groups including Bacillus sp., Corynebacterium sp., Pseudomonas sp., E. coli, Bacillus sp., Yersinia sp., Planococcus sp., Yersinia sp., Klebsiella sp., Agrobacterium sp. The rusted acrylic wall sample was found with Corynebacterium sp., Flavobacterium sp., Neisseria sp., Staphylococcus sp., Streptococcus sp., Pseudomonas sp., Bacillus sp. and Klebseilla sp.
The defining features of the isolates have shown that from the eighteen isolates, nine isolates belong to the Gram-positive and the remaining nine belong to the Gram-negative group (Table 3-4). The catalase test showed positive in all the bacterial isolates and the carbohydrate fermentation test also showed positive in all the bacterial isolates except for the only one isolated from the rusted acrylic wall paint sample. The Protease test and Voges Proskauer test were negative in all isolates of the two experimental sites.
|
| Table 3: | Biochemical analysis of isolated bacterial strains, isolated from the rusted acrylic wall paint | |||
| Bacterial isolate identification code |
Colony colour |
Shape of the bacteria |
Gram staining |
Catalase test |
Carbohydrate test (Maltose) |
Gas production |
Starch hydrolase test |
Motility test |
Indole test |
H2S production |
VP test |
Methyl red |
Citrate test |
Protease test |
Identified as |
| A | White |
Rod |
+ |
+ |
+ |
- |
- |
+ |
- |
- |
- |
+ |
- |
- |
Bacillus sp. |
| B | Yellow |
Rod |
+ |
+ |
+ |
- |
- |
+ |
- |
- |
- |
- |
- |
- |
Corynebacterium sp. |
| C | Pink |
Rod |
- |
+ |
+ |
- |
- |
+ |
- |
- |
- |
+ |
- |
- |
Pseudomanas sp. |
| D | White |
Rod |
- |
+ |
+ |
- |
- |
+ |
- |
- |
- |
+ |
- |
- |
Escherichia coli |
| E | Yellow |
Rod |
+ |
+ |
+ |
- |
- |
+ |
- |
- |
- |
+ |
- |
- |
Bacillus sp. |
| F | White |
Cocci |
- |
+ |
+ |
- |
- |
+ |
- |
- |
- |
+ |
- |
- |
Yersinia sp. |
| G | Yellow |
Cocci |
+ |
+ |
+ |
- |
- |
+ |
- |
- |
- |
+ |
- |
- |
Planococcus sp. |
| H | White |
Cocci |
- |
+ |
+ |
- |
- |
+ |
- |
- |
- |
+ |
+ |
- |
Yersinia sp. |
| I | White |
Rod |
- |
+ |
+ |
+ |
- |
+ |
- |
- |
- |
- |
+ |
- |
Klebsiella sp. |
| J | Yellow |
Rod |
- |
+ |
+ |
+ |
- |
+ |
- |
- |
- |
+ |
- |
- |
Agrobacterium sp. |
| +: Indicates positive result and -: Indicates the negative result of the particular bacterial species to the mentioned biochemical tests | |||||||||||||||
| Table 4: | Biochemical analysis of isolated bacterial strains, isolated from the rusted acrylic wall paint | |||
| Bacterial isolate identification code |
Colony colour |
Shape of the bacteria |
Gram staining |
Catalase test |
Carbohydrate test (Maltose) |
Gas production |
Starch hydrolase test |
Motility test |
Indole test |
H2S production |
VP test |
Methyl red |
Citrate test |
Protease test |
Identified as |
| K | White |
Rod |
+ |
+ |
+ |
- |
- |
Non motile |
- |
+ |
- |
+ |
- |
- |
Corynebacterium sp. |
| L | Yellow |
Rod |
+ |
+ |
+ |
- |
- |
Motile |
- |
+ |
- |
+ |
- |
- |
Flavobacterium sp. |
| M | Pale yellow |
Cocci |
- |
+ |
+ |
- |
- |
Motile |
- |
- |
- |
+ |
- |
- |
Neisseria sp. |
| N | White |
Cocci |
+ |
+ |
+ |
- |
- |
Motile |
- |
- |
- |
+ |
+ |
- |
Staphylococcus sp. |
| O | White |
Cocci |
+ |
- |
+ |
- |
- |
Non motile |
+ |
- |
- |
+ |
- |
- |
Streptococcus sp. |
| P | Pale yellow |
Rod |
- |
+ |
- |
+ |
- |
Motile |
- |
- |
- |
- |
- |
- |
Pseudomonas sp. |
| Q | Yellow |
Rod |
+ |
+ |
+ |
- |
- |
Motile |
- |
- |
- |
+ |
- |
- |
Bacillus sp. |
| R | White |
Rod |
- |
+ |
+ |
- |
- |
Non motile |
+ |
- |
- |
+ |
- |
- |
Klebsiella sp. |
| +: Indicates positive result and -: Indicates the negative result of the particular bacterial species to the +mentioned biochemical tests respectively | |||||||||||||||
On the contrary, the methyl red test showed positive in all isolates except three isolates. The isolated species belong to twelve genera. A total of eighteen bacterial species were isolated which include Corynebacterium sp., Flavobacterium sp., Neisseria sp., Staphylococcus sp., Streptococcus sp., Pseudomonas sp., Bacillus sp., Klebsiella sp., Bacillus sp., Corynebacterium sp., Pseudomonas sp., E. coli, Bacillus sp., Yersinia sp., Planococcus sp., Yersinia sp., Klebsiella sp. and Agrobacterium sp. genera like Bacillus, Corynebacterium, Pseudomonas and Klebsiella. These were identified using the colony morphology and biochemical tests, performed in the soil samples, collected from the experimental sites.
DISCUSSION
The results of the study revealed the presence of some of the major microbes inhabiting species of contaminated sites which including Bacillus, Corynebacterium, Flavobacterium, Neisseria, Staphylococcus, Streptococcus, Pseudomonas, Escherichia, Yersinia, Planococcus, Agrobacterium and Klebsiella. (Table 3). Several reports have proven the presence of bacterial isolates in plastic-contaminated soils, has potentially eliminating the pollutants24,25. Mukherjee and Chatterjee26 introspected the comparative extent of plastic biodegradation by the application of Aspergillus niger and three bacterial species of Bacillus weihenstephanensis, Burkholderia cepacia and Escherichia coli which were isolated from hydrocarbon-enriched soil. Similarly, Lactobacillus sp. was isolated and identified from the rusted acrylic wall paint samples. The biodegradation potency of the following strains such as Bacillus cereus, B. subtilis, B. megaterium, Brevibacillus borstelensis have been evaluated27.
The isolates consuming the Low Density Polythene (LDPE) material, as a carbon source by the B. cereus, B. subtilis and B. megaterium were detected using the standard characterization protocols28. Both bacteria, especially, Pseudomonas, Bacillus, Brevibacillus, Cellulosimicrobium, Lysinibacillus and Aspergillus fungus were implicated in polyethylene degrading role in the plastic dumpsite28. The isolate of Pseudomonas species from the long-term plastic waste land-fill area, would be employed in the biodegradation of plastic waste materials.
The formation of biofilm by the bacteria, followed by the reduced durability of polymer structures would be beneficial in the plastic degradation studies. Bacteria can be present on an apparently clean surface in sufficient numbers to exert adverse corrosion effects over the concrete and metal surfaces, due to the production of inorganic substances29. Chemolithotrophic and oligotrophic bacteria were found to be colonizing at very low nutrient levels, leading them more amenable to colonization by other microorganisms30. Likewise, various bacterial isolates were allured from the rusted acrylic paint namely Corynebacterium sp., Flavobacterium sp., Neisseria sp., Staphylococcus sp., Streptococcus sp., Pseudomonas sp., Bacillus sp. and Klebsiella sp.
Gram-negative bacteria were predominantly encountered among all the bacterial isolates when compared with the gram-positive group. Pseudomonas and Enterobacter are opportunistic pathogens, often found to be present in the landfills of waste materials31, further supports the results of this experiment. Correspondingly, in this present study, gram-positive and gram-negative bacteria were equally distributed in both contaminated zones. In plastic-contaminated sites, gram-negative bacteria were higher in proportion and in rusted acrylic wall paint, gram-positive were higher in proportion.
Therefore, essential means of monitoring the plastic landfill and acrylic polymer rust becomes inevitable for the enumeration of the microbial community and further to devise the mechanism to adapt the proper management principles and technologies for improved waste management. The present study results on the bacterial isolates might have the ability and the experimental results have a further scope on the experimenting to test the degradability through the intracellular and extracellular depolymerase enzymes to oxidize the complex polymers into simpler compounds, as some of the studies are available to show the possible bio-degradation of plastic wastes32.
The experimental results have practical implications by providing information on bacterial communities and their functional biochemical metabolism for plastic polymer degradation and would contribute towards a clear insight into these aspects. However, there is a wider scope is available to extend this study further, to find out the bacteria and other microbial organisms in the various plastic wastes environment. This comprehensive information will help in the microbial degradation of synthetic polymers towards mitigation of plastic waste and to gain knowledge in the development of sustainable plastic waste management guidelines in the future.
CONCLUSION
The present study delivers insights into the bacterial community of the contaminated sites of plastic substances and their possible role in the degradation of synthetic polyethylene substances. A number of plastic degrading bacteria have been sourced from the adverse environmental polluted conditions including long-term plastic waste landfills and the leaked and rusted exterior wall, selected in this experiment. Ten different bacterial species and eight bacterial species were isolated respectively from the soil samples collected from the long-term plastic waste filling site and the rusted wall, due to water pipe leakage. These microbial populations would have the potential in the plastic degradation and the study results can be applied to extend the work to test the biodegradation of plastic wastes. Further, there is an ample scope in the isolation of fungi and other microbial populations using the experimental approach.
SIGNIFICANCE STATEMENT
This experiment results demonstrate the occurrence of bacterial organisms from the long-term plastic waste landfill site and over the rusted exterior walls with acrylic degradation. The existence of the bacteria was found in the two experimental sites, prone to be contaminated and found that the bacterial organisms are able to adapt in such heavily contaminated site conditions. Degradation of plastic waste using the bacterial species, which can adapt in this environment, would be beneficial in the contribution towards the plastic pollution management.
REFERENCES
- Hopewell, J., R. Dvorak and E. Kosior, 2009. Plastics recycling: Challenges and opportunities. Philos. Trans. Royal Soc. B. Biol. Sci., 364: 2115-2126.
- Okunola, A.A., K.I. Ologbonjaye, O. Awosolu and O.E. Alalade, 2019. Public and environmental health effects of plastic wastes disposal: A review. J. Toxicol. Risk Assess., 5.
- Oehlmann, J., U. Schulte-Oehlmann, W. Kloas, O. Jagnytsch and I. Lutz et al., 2009. A critical analysis of the biological impacts of plasticizers on wildlife. Phil. Trans. R. Soc. B, 364: 2047-2062.
- Zettler, E.R., T.J. Mincer and L.A. Amaral-Zettler, 2013. Life in the “Plastisphere”: Microbial communities on plastic marine debris. Environ. Sci. Technol., 47: 7137-7146.
- de Tender, C.A., L.I. Devriese, A. Haegeman, S. Maes, T. Ruttink and P. Dawyndt, 2015. Bacterial community profiling of plastic litter in the Belgian part of the North Sea. Environ. Sci. Technol., 49: 9629-9638.
- Munir, E., R.S.M. Harefa, N. Priyani and D. Suryanto, 2018. Plastic degrading fungi Trichoderma viride and Aspergillus nomius isolated from local landfill soil in Medan. IOP Conf. Ser.: Earth Environ. Sci., 126.
- Du, Y., X. Liu, X. Dong and Z. Yin, 2022. A review on marine plastisphere: Biodiversity, formation, and role in degradation. Comput. Struct. Biotechnol. J., 20: 975-988.
- Vaksmaa, A., K. Knittel, A.A. Asbun, M. Goudriaan and A. Ellrott et al., 2021. Microbial communities on plastic polymers in the Mediterranean Sea. Front. Microbiol., 12.
- Basili, M., G.M. Quero, D. Giovannelli, E. Manini and C. Vignaroli et al., 2020. Major role of surrounding environment in shaping biofilm community composition on marine plastic debris. Front. Mar. Sci., 7.
- Malik, S., M. Beer, M. Megharaj and R. Naidu, 2008. The use of molecular techniques to characterize the microbial communities in contaminated soil and water. Environ. Int., 34: 265-276.
- Jorquera, M.A., S.P. Graether and F. Maruyama, 2019. Editorial: Bioprospecting and biotechnology of extremophiles. Front. Bioeng. Biotechnol., 7.
- Tremblay, J., E. Yergeau, N. Fortin, S. Cobanli and M. Elias et al., 2017. Chemical dispersants enhance the activity of oil- and gas condensate-degrading marine bacteria. ISME J., 11: 2793-2808.
- Margesin, R., D. Labbé, F. Schinner, C.W. Greer and L.G. Whyte, 2003. Characterization of hydrocarbon-degrading microbial populations in contaminated and pristine alpine soils. Appl. Environ. Microbiol., 69: 3085-3092.
- Chaerun, S.K., K. Tazaki, R. Asada and K. Kogure, 2004. Bioremediation of coastal areas 5 years after the Nakhodka oil spill in the Sea of Japan: Isolation and characterization of hydrocarbon-degrading bacteria. Environ. Int., 30: 911-922.
- Jin, H.M., J.M. Kim, H.J. Lee, E.L. Madsen and C.O. Jeon, 2012. Alteromonas as a key agent of polycyclic aromatic hydrocarbon biodegradation in crude oil-contaminated coastal sediment. Environ. Sci. Technol., 46: 7731-7740.
- Varjani, S.J. and V.N. Upasani, 2016. Biodegradation of petroleum hydrocarbons by oleophilic strain of Pseudomonas aeruginosa NCIM 5514. Bioresour. Technol., 222: 195-201.
- Sarkar, P., A. Roy, S. Pal, B. Mohapatra, S.K. Kazy, M.K. Maiti and P. Sar, 2017. Enrichment and characterization of hydrocarbon-degrading bacteria from petroleum refinery waste as potent bioaugmentation agent for in situ bioremediation. Bioresour. Technol., 242: 15-27.
- Varjani, S.J., 2017. Microbial degradation of petroleum hydrocarbons. Bioresour. Technol., 223: 277-286.
- Xu, X., Z. Zhai, H. Li, Q. Wang, X. Han and H. Yu, 2017. Synergetic effect of bio-photocatalytic hybrid system: g-C3N4 and Acinetobacter sp. JLS1 for enhanced degradation of C16 alkane. Chem. Eng. J., 323: 520-529.
- Sekiguchi, T., T. Sato, M. Enoki, H. Kanehiro, K. Uematsu and C. Kato, 2011. Isolation and characterization of biodegradable plastic degrading bacteria from deep-sea environments. JAMSTEC Rep. Res. Dev., 11: 33-41.
- Gómez-Alarcón, G., M.L. Muñoz and M. Flores, 1994. Excretion of organic acids by fungal strains isolated from decayed sandstone. Int. Biodeterior. Biodegrad., 34: 169-180.
- Sani, Z.M., A.S. Dalhatu and S. Ibrahim, 2021. Comparative study of the potentials of Aspergillus terreus, Bacillus species and Chlorella vulgaris on the bio-remediation of reactive red 198 (RR198) dye. UJMR: J. Microbiol. Res., 6: 168-174.
- Brenner, D.J., J.T. Staley and N.R. Krieg, 2005. Classification of Procaryotic Organisms and the Concept of Bacterial Speciation. In: Bergey's Manual® of Systematic Bacteriology, Brenner, D.J., N.R. Krieg, J.T. Staley and G.M. Garrity (Eds.), Springer, Boston, Massachusetts, US, ISBN: 978-0-387-24143-2, pp: 27-33.
- Gupta, S.B., A. Ghosh and T. Chowdhury, 2010. Isolation and selection of stress tolerant plastic loving bacterial isolates from old plastic wastes. World J. Agric. Sci., 6: 138-140.
- Usha, R., T. Sangeetha and M. Palaniswamy, 2011. Screening of polyethylene degrading microorganisms from garbage soil. Libyan Agric. Res. Center J. Int., 2: 200-204.
- Mukherjee, S. and S. Chatterjee, 2014. A comparative study of commercially available plastic carry bag biodegradation by microorganisms isolated from hydrocarbon effluent enriched soil. Int. J. Curr. Microbiol. Appl. Sci., 3: 318-325.
- Abrusci, C., J.L. Pablos, T. Corrales, J. López-Marín, I. Marín and F. Catalina, 2011. Biodegradation of photo-degraded mulching films based on polyethylenes and stearates of calcium and iron as pro-oxidant additives. Int. Biodeterior. Biodegrad., 65: 451-459.
- Muhonja, C.N., H. Makonde, G. Magoma and M. Imbuga, 2018. Biodegradability of polyethylene by bacteria and fungi from Dandora dumpsite Nairobi-Kenya. PLoS ONE, 13.
- Ravikumar, H.R., S.S. Rao and C.S. Karigar, 2012. Biodegradation of paints: A current status. Indian J. Sci. Technol., 5: 1977-1987.
- Scheerer, S., O. Ortega-Morales and C. Gaylarde, 2009. Microbial Deterioration of Stone Monuments-An Updated Overview. In: Advances in Applied Microbiology, Laskin, A.I., S. Sariaslani and G.M. Gadd (Eds.), Academic Press, Cambridge, Massachusetts, ISBN: 9780123747884, pp: 97-139.
- Munir, E., D. Suryanto, Y. Pasaribu, S. Mubtasima, A. Hartanto, A. Lutfia and A.F. Nasution, 2022. Occurrence of microbial community on plastic wastes in Terjun Landfill, North Sumatra. IOP Conf. Ser.: Earth Environ. Sci., 1115.
- Haedar, N., T. Clara, Fahrudin, A. Abdullah, S. Fausiah and M.T. Rapak, 2019. Selection of plastic degradation indigenous bacteria isolated from Tamangapa Landfill Macassar city. J. Phys.: Conf. Ser., 1341.
How to Cite this paper?
APA-7 Style
Subramanian,
V., Ramamoorthi,
P., Pandian,
K.D. (2024). Bacterial Communities in Plastic-Contaminated Sites and Rusted Acrylic Wall Paint: Isolation and Characterization. Bacteriology Journal, 14(1), 1-9. https://doi.org/10.3923/bj.2024.1.9
ACS Style
Subramanian,
V.; Ramamoorthi,
P.; Pandian,
K.D. Bacterial Communities in Plastic-Contaminated Sites and Rusted Acrylic Wall Paint: Isolation and Characterization. Bacteriol. J 2024, 14, 1-9. https://doi.org/10.3923/bj.2024.1.9
AMA Style
Subramanian
V, Ramamoorthi
P, Pandian
KD. Bacterial Communities in Plastic-Contaminated Sites and Rusted Acrylic Wall Paint: Isolation and Characterization. Bacteriology Journal. 2024; 14(1): 1-9. https://doi.org/10.3923/bj.2024.1.9
Chicago/Turabian Style
Subramanian, Vishalakshi, Priyanka Ramamoorthi, and Kannan Dorai Pandian.
2024. "Bacterial Communities in Plastic-Contaminated Sites and Rusted Acrylic Wall Paint: Isolation and Characterization" Bacteriology Journal 14, no. 1: 1-9. https://doi.org/10.3923/bj.2024.1.9

This work is licensed under a Creative Commons Attribution 4.0 International License.



